Posted by Osvaldo
September 7, 2016
One of the nice things of my job is the close contact between the experiments and the analysis. While my collaborators develop the method in the lab, working with reagents, patients samples and sophisticated machines, I sit in front of a computer developing tools like this and using it to analyse the results coming from the lab.
Recently, one interesting question was raised when a student defended her PhD thesis. I wasn’t there so I was only asked about this later. The question was “did the development of the experimental method and that of the analysis pipeline influence each other?” No, said one colleague. Of course they did, I later replied.
Why did we have different opinions, since we both know how everything was developed? My reasoning was that, without a computational pipeline to analyse the results produced in the lab, my colleagues would have had hardly any hint on whether their experiments were working or not. At the same time I could test my tools on data of realistic size, with realistic signal-to-noise ratio and so on. One side could not have done it without the other. My colleague, maybe because of the context in which the question was asked, intended it in a more specific way: did you tweak the parameters in the software in order to specifically respond to a change in the experimental conditions? And the answer here is no, undoubtedly.
My most successful tool, at least in terms of citations, is a software to denoise DNA sequences, chiefly in the analysis of viral samples. It is an unsupervised learning algorithm that models in an explicit way, though simplified, the process of sequencing a genetically diverse sample. We first wrote the generative model, then the algorithm to do inference on its parameters and we implemented it in an efficient way. There is a nice correspondence between parameters in the model and physical processes taking place in the lab. Thus, it is easily interpretable.
It was great, but it took quite some time.
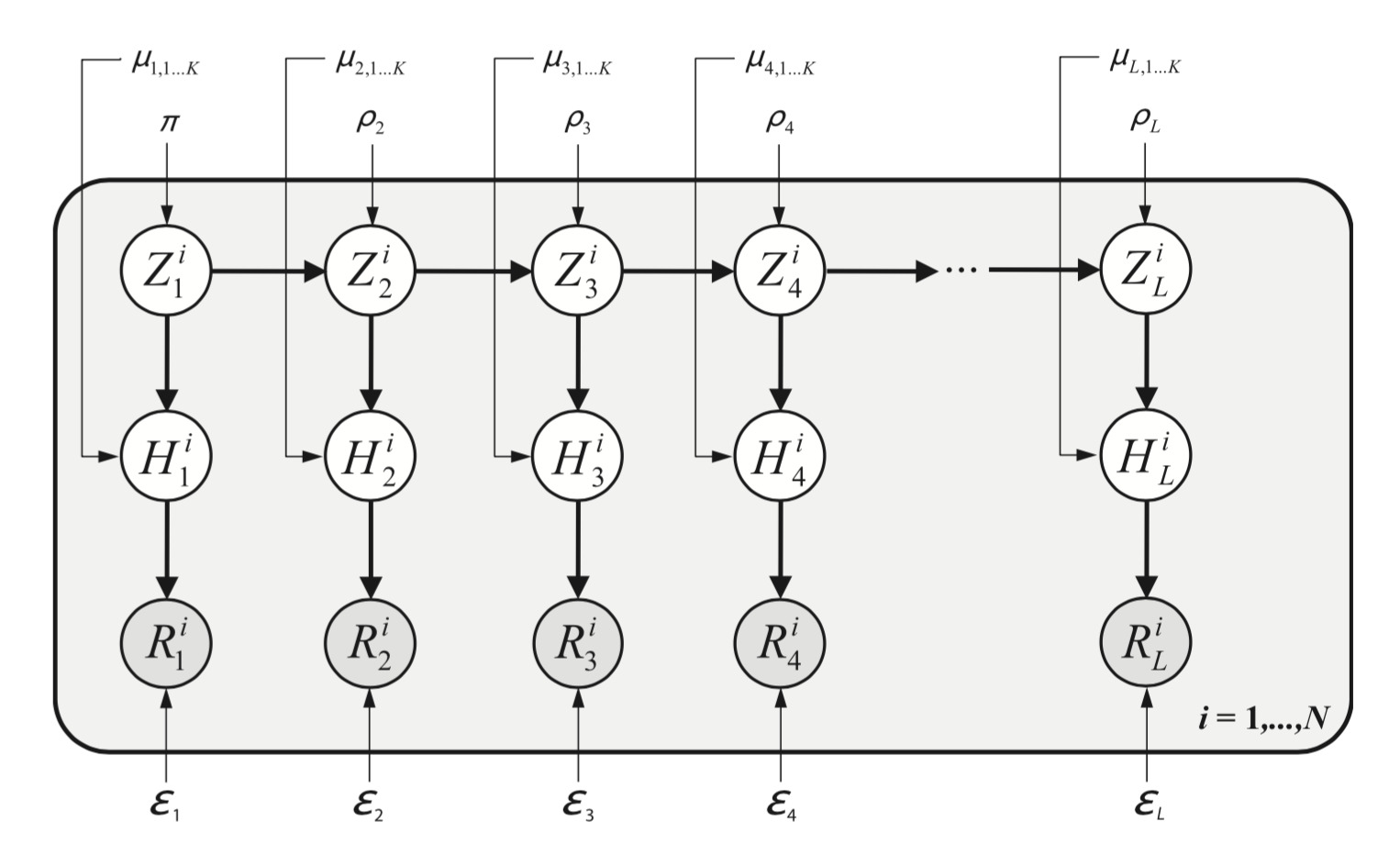
Graphical representation of a generative model
But do you always need such a model? According to some, the goal of a model is to provide a simple low-dimensional summary of a dataset
, and I would stretch this to say that even a simple data visualisation can be considered a model.
For example, in this more recent project I relied heavily on some heuristic tools to denoise my data, match them by similarity and obtain summary statistics. Advantages of these heruristic tools: they are fast, thoroughly tested by a whole community of researchers, and I could easily use them. I did not have to struggle to write efficient code from scratch. On the other hand, I do not have many parameters that I could directly link to anything that is happening in the lab.
A slightly related topic of discussion in machine learning is the trade-off between interpretability and accuracy of a model. Although accuracy here depends on many more things, this observation makes me appreciate even more the power of some methods in machine learning. The models underlying tools like clustering, trees, regression are so general and powerful that they can be routinely used for a plethora of problems in all sorts of domains with great results. It maybe worth reminding it next time we apply one of these.
Header image from Flickr